The Eastern side Armenians
Hovhannisyan et al. 2024 published numerous DNA from Artsakh, Syunik and Gardman regions. Additionally a member of our group Diana Sarkisova Vikutan provided a number of coordinates from Shamakhi Armenians, as well as DNA with mixed ancestry from Shamakhi and Artsakh regions. We are grateful to her for this action.
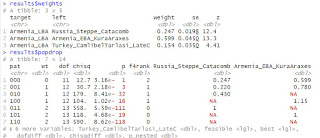
+ Artsakh Armenians are similar to Syunik Armenians. Both are part of the genetic cline of historic Armenia which have strong affinity to so called "Central farmer" ancestry. Gardman Armenians also are close to them with some Caucasian hunter gatherer (CHG) shift.
+ All Armenian subgroups are away from Armenia LIA samples from Syunik. This is due to so called "southern" shift which might have occurred in those eastern regions some time after Urartu.
+ Shamakhi Armenians have higher steppe ancestry than any known Armenian subgroup. Two samples from Shamakhi region are somewhat different from each other. One of them Sham01 has more Zagros Neo ancestry. What is the source of this shift is at this stage unknown. It must be noted that a single Late Antiquity (3td century AD) sample from Shamakhi region had already an extra Zagros/Iran Neolithic ancestry almost certainly due to Iranian influence on that region. Another possibility is that the Zagros shift in Sham01 is from a more modern non-Armenian ethnic group. And another possibility is that yet unsampled mysterious Tat Armenians subgroup is the origin of this extra shift.
+ Three DNA samples have a mix Shamakhi and Artsakh ancestry. They are between the Shamakhi and Artsakh points as expected. Nevertheless their dispersal means that the real diversity of Shamakhi Armenians could well be as high as already available two samples himts.
+ And finally Udi people are far from their linguistic kinsmen Lezgins. Even more they are shifted away from Late Antiquity Shamakhi sample who might represent the Caucasian Albanian ancestry. How Udi genetic profile formed is unknown yet but their proximity to Armenians who themselves are shifted toward "south-west" means that Udi got Armenian ancestry either in antiquity or more likely in medieval period. It must be noted that some Azerbaijani from Shaki region also plot close to Udi meaning that the impact of Armenian like ancestry was not restricted just to Udi.
Additionally the Skourtanioti 2024 paper noticed an"Anatolian" shift in Iberia and linked it to the spread of Christianity. It's possible that similar events occurred in Caucasian Albania in Late Antiquity and Early Medieval period. Hopefully with the publication of time transection from Kakhetia region we can learn better how the genetics of Caucasian Albania evolved in historic times.
Summing up this results we can say that the genetic history of what is now the Azerbaijani Republic is quite complex after Bronze and Iron Age. Multiple influences shaped the genetic diversity of the region. The single Caucasian Albanian sample is somewhat puzzling. Do it represent a regular Albanian ancestry or was an outlier is unknown. As usual more modern and ancient samples are needed from those regions to better understand it's history.
If You are an Armenian and have ancestry from Gardman, Shaki and Shirvan including Shamakhi regions You can join the relevant FTDNA project. You can find the link below. The title of the project will change in the future.